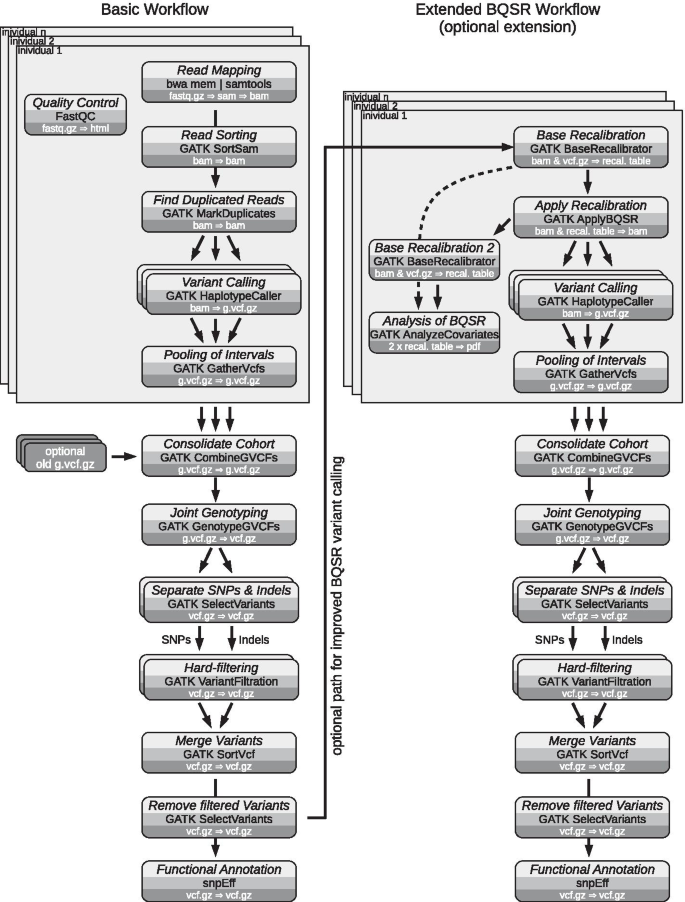
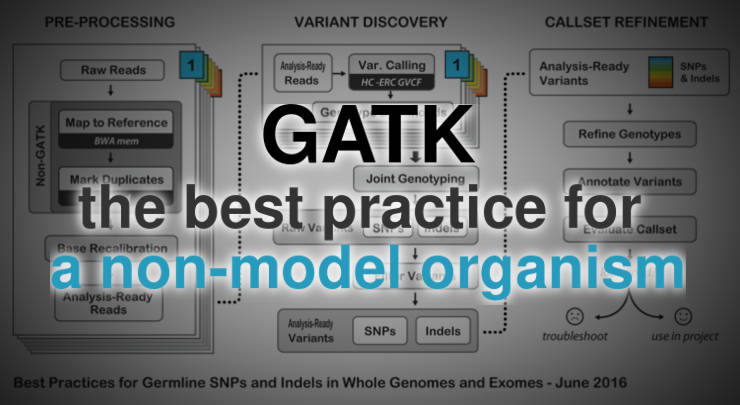
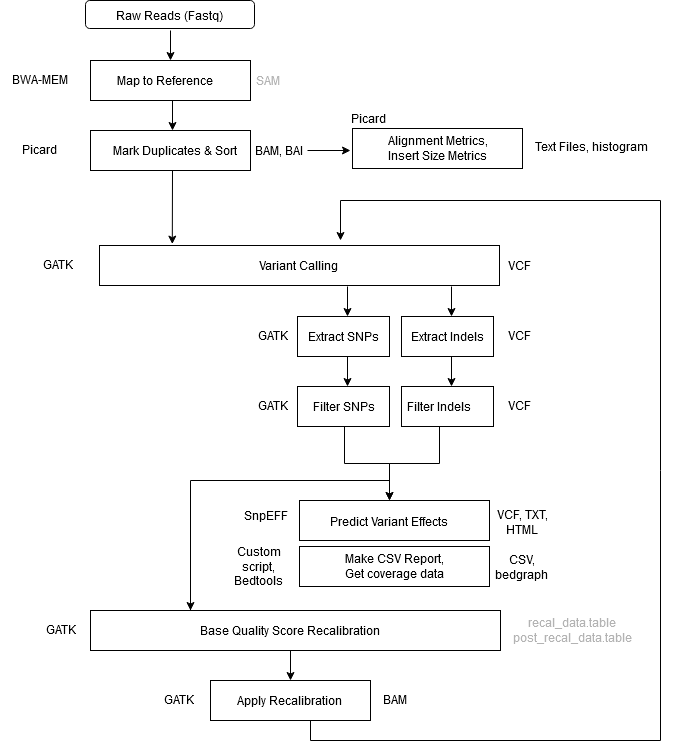
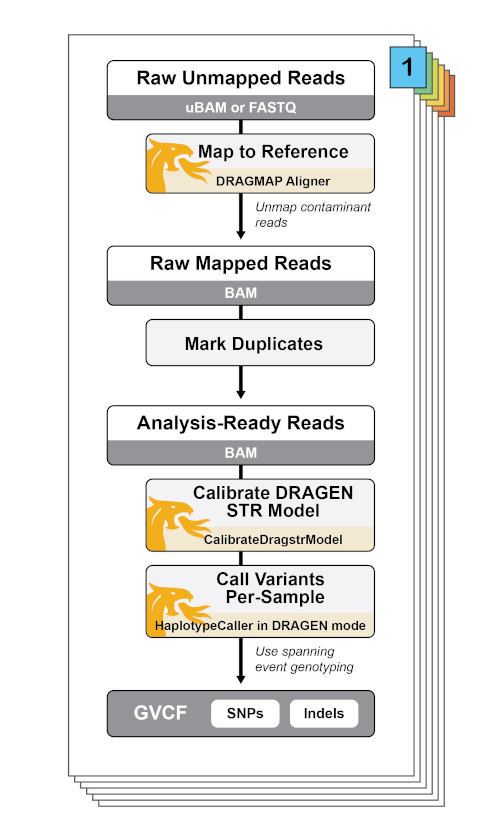
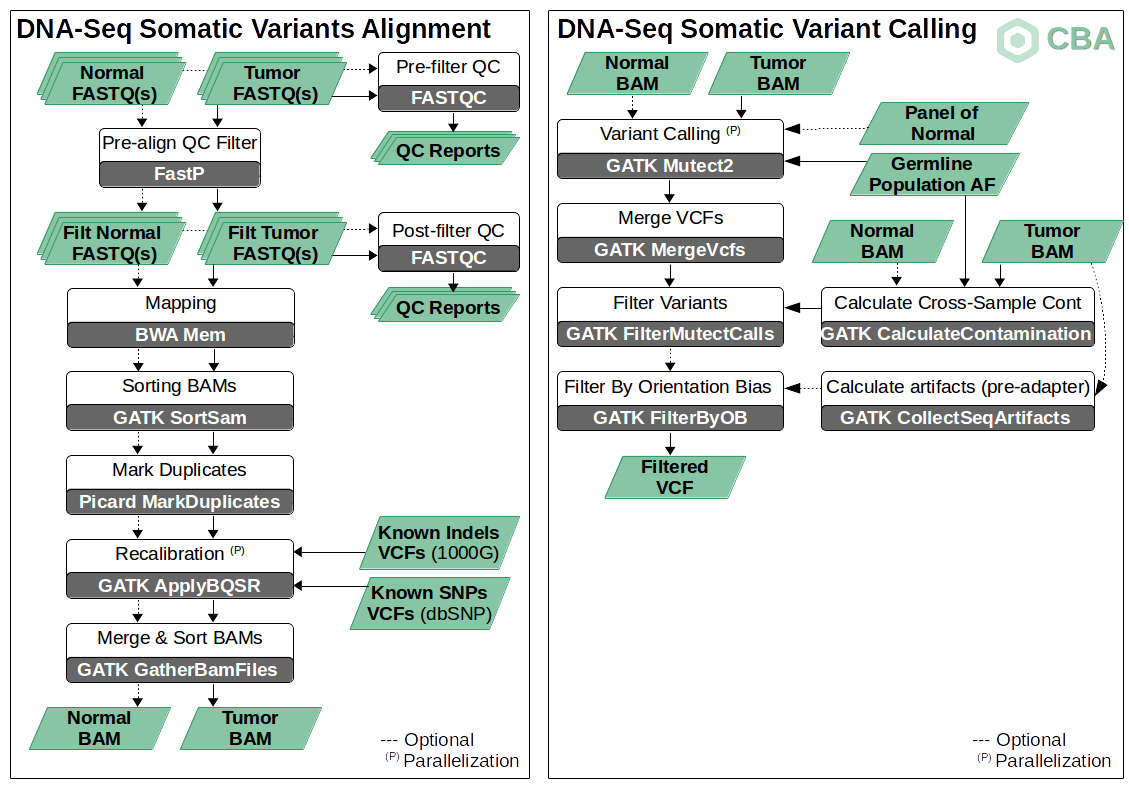
Protocol for unbiased, consolidated variant calling from whole exome sequencing data - ScienceDirect
GitHub - crukci-bioinformatics/gatk-tools: Utilities for processing sequencing data and genomic variants using GATK

Amazon.fr - Genomics in the Cloud: Using Docker, GATK, and WDL in Terra - Auwera, Geraldine van der, O'Connor, Brian D. - Livres

Broad Institute to release Genome Analysis Toolkit 4 (GATK4) as open source resource to accelerate research | Broad Institute
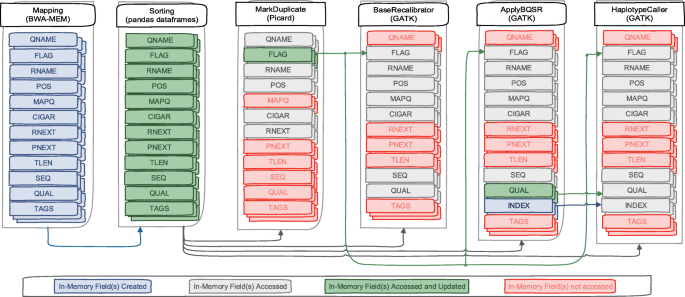
Optimizing performance of GATK workflows using Apache Arrow In-Memory data framework | BMC Genomics | Full Text

Comparison of calling pipelines for whole genome sequencing: an empirical study demonstrating the importance of mapping and alignment | Scientific Reports